VIPpred: A novel tool for predicting variant impact on phosphorylation (VIP) events driving carcinogenesis |
|||||||||||
|
Disrupted protein phosphorylation due to genetic variation is a widespread phenomenon that triggers oncogenic transformation of healthy cells. However, few relevant phosphorylation disruption events have been verified due to limited biological experimental methods. Because of the lack of reliable benchmark datasets, current bioinformatics methods primarily use sequence-based traits to study variant impact on phosphorylation (VIP). Here, we developed a novel tool called VIPpred that allowed us to successfully identify different types of variant impact on phosphorylation (VIP) events based on the first high-throughput VIP dataset with spectral evidence generated by VIPpipeline. VIPpred serves as a machine learning method characterized by multidimensional features that exhibits robust performance across different cancer types. We also provide a pan-cancer landscape of VIP events based on genetic variants derived from TCGA Pan-Cancer Atlas project. Furthermore, our newly developed VIP analysis process has the potential to be extended to other non-cancer disease and predicting changes of other modification status in cell regulatory mechanism. |
|
||||||||||
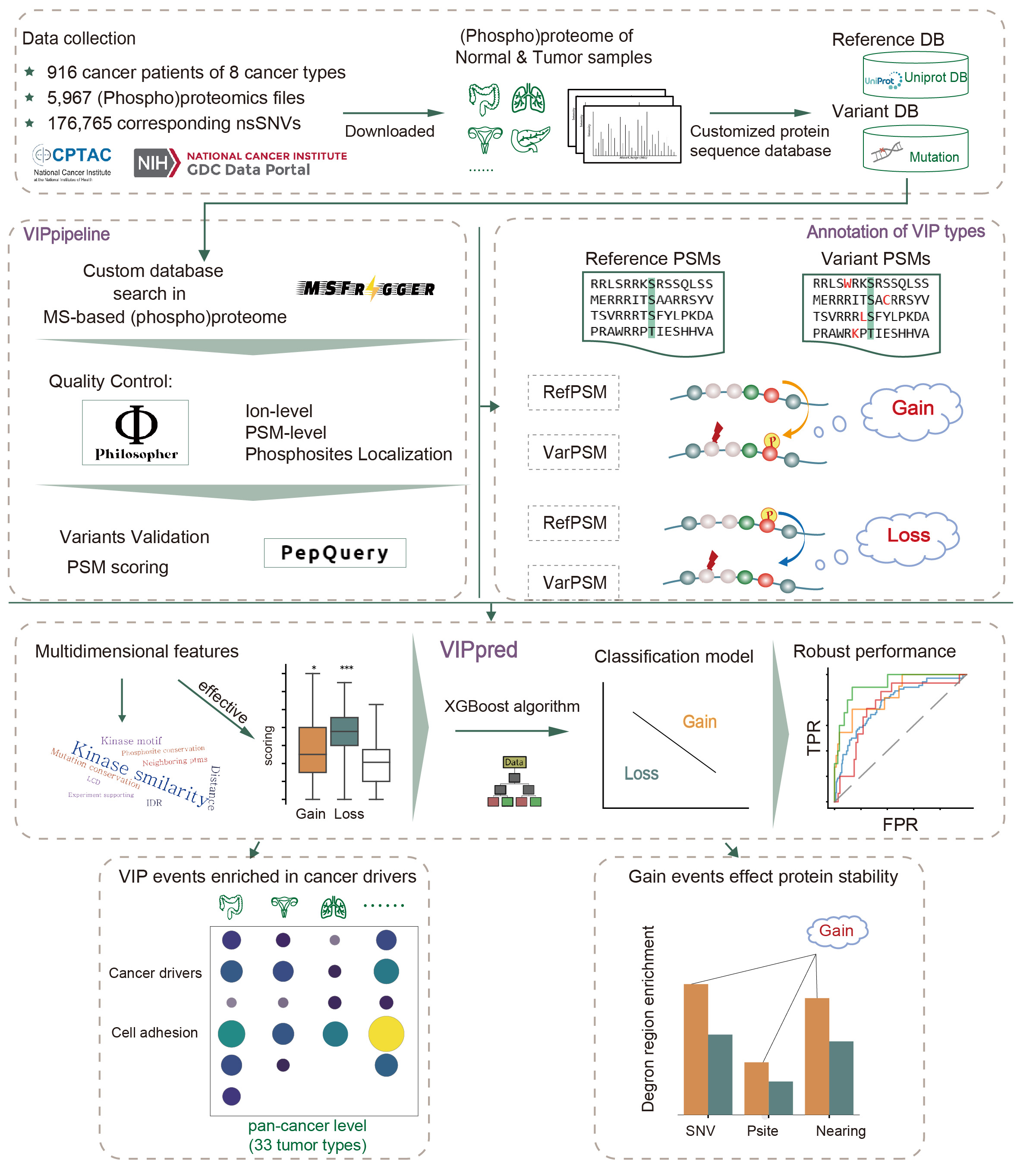
|
Cite: Xiaofeng Xu, Ying Li, Taoyu Chen, Chao Hou, Liang Yang, Peiyu Zhu, Yi Zhang, Tingting Li. VIPpred: A novel tool for predicting variant impact on phosphorylation (VIP) events driving carcinogenesis. Contact Us VIPpred is free for non-commercial use for academic, government and non-profit institutions. For
commercial
use, questions or suggestions.
please contact the authors. |
||||||||||